
Dept
of
Animal
Physiol
Group
re-
search
page |
CELL PROLIFERATION
GROUP
The role of the polyamines in
cell cycle control and programmed cell
death
General
background |
|
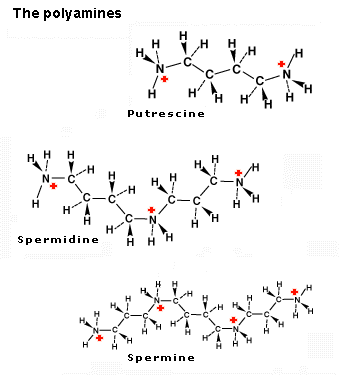
The polyamines - putrescine, spermidine, and
spermine - constitute a group of cell components that are
important in the regulation of cell proliferation and cell
differentiation. There is also evidence suggesting a role for
polyamines in programmed cell death. Although their exact functions
have not yet been identified, it is clear that the polyamines play
important roles in a number of cellular processes such as
replication, transcription, and translation. Presumably these roles
are exerted by specific interactions that can only be mediated by
the cationic polyamines with their characteristic, unique, and
flexible charge distributions (see figure below).
|
|
The importance of the polyamines in cell function is reflected in
a strict regulatory control of their intracellular levels. Adequate
cellular polyamine levels are achieved by a careful balance between
biosynthesis, degradation, and uptake of the amines. Some of the
regulatory mechanisms involved in maintaining a balance in the
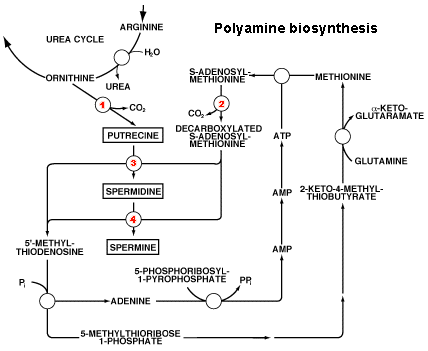
cellular polyamine pools are truly unique. The polyamine
biosynthetic pathway consists of two highly-regulated enzymes,
ornithine decarboxylase (1 in the
figure below) and S-adenosylmethionine decarboxylase (2 in the figure below), and two
constitutively expressed enzymes, spermidine synthase (3 in the figure below) and spermine
synthase (4 in the figure below).
The biological half-lifes of the two regulatory enzymes ornithine
decarboxylase and S-adenosylmethionine decarboxylase (5-60 min) are
among the shortest known for mammalian enzymes, allowing the cell to
rapidly change the cellular polyamine levels.
|
|
The polyamine degradation pathway consists of the
highly-regulated enzyme spermidine/spermine N1-acetyl transferase
and the constitutively expressed polyamine oxidase. In addition to
regulation of polyamine levels by biosynthesis and degradation,
cells are equipped with an efficient transport system for
utilization of exogenously derived polyamines.
The
biosynthesis of polyamines is increased by a great variety of
physiological growth stimuli, and polyamine deficiency results in an
arrest of cell proliferation, which can be reversed by
supplementation with external polyamines. Polyamine deficiency may
be achieved by treating cells with specific inhibitors of the
polyamine biosynthetic enzymes. In view of the fact that
constitutive overproduction of ornithine decarboxylase has been
observed in many types of cancer cells, the ornithine decarboxylase
gene appears to be of central importance in the regulation of cell
growth. When the ornithine decarboxylase gene is transfected into
cells and overexpressed, the cells go through malignant
transformation.
Most studies concerning the regulation of
polyamine biosynthesis and the function of the polyamines have been
performed with asynchronous cell systems. Thus, results obtained are
mean values depending on the cell cycle phase distributions of the
cells and the variations of the parameter studied during the cell
cycle. To overcome this technical problem, we have focused on using
cell populations that progress synchronously and unperturbed through
the cell cycle. In Chinese hamster ovary cells progressing
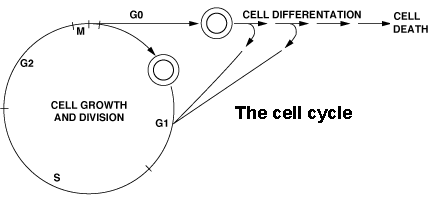
synchronously through the cell cycle, we found that polyamine
biosynthesis was activated at the G1 to S and G2 to M transitions.
In addition, we found that ornithine decarboxylase and
S-adenosylmethionine decarboxylase are regulated at both
transcriptional and translational levels during the cell
cycle. |
|
Further work is needed, before we clearly understand how
polyamine biosynthesis is regulated during the cell cycle. Such work
would provide a background frame helping us to understand the role
of the polyamines in cell cycle regulation (which is already known
to be dependent on oncogenes, tumor suppressor genes, and cyclins
with their associated kinases).
The fact that polyamine
biosynthesis was activated at the G1 to S transition infers a role
for the polyamines in DNA replication. Our earlier results have
shown that polyamine depletion affected DNA replication negatively,
presumably by reducing the rate of DNA elongation. Recently we
presented evidence that polyamine biosynthesis inhibition affects
DNA replication in Chinese hamster ovary cells within one cell cycle
after seeding them in the presence of polyamine synthesis inhibitors
(either ornithine decarboxylase or S-adenosylmethionine
decarboxylase inhibitors). The G1 to S transition and the G2 phase
progression were affected only after several cell
cycles.
Staining of bromodeoxyuridine labelled cells with
fluorescently-labelled antibodies against bromodeoxyuridine (which
is a thymidine analog) has demonstrated that eukaryotic DNA
replication is concentrated to discrete foci in the interphase
nucleus. The number and the spatial distribution of replication
domains vary in a specific pattern throughout the S phase. Using
confocal laser scanning microscopy, we found that neither the number
of replication domains nor their spatial distribution was affected
in polyamine-depleted cells. This would have been the case if DNA
replication initiation was affected. Further work is needed to
establish whether the polyamines have a specific role in DNA
replication.
DNA replication requires the presence of a
number of proteins which have a direct or indirect role for the
incorporation of the nucleotides into DNA as well as for the
structural integrity of the chromatin. Topoisomerases are required
to release various stresses in DNA that might affect the progress of
the replication machinery. Using a decatenation assay, we have found
that the topoisomerase II activity in cell extracts is the same in
control and polyamine-depleted cells. However, when using the single
cell gel electrophoresis assay to study the actual efficiency of
topoisomerase II in situ, we found that the enzyme was less
active in polyamine-depleted cells than in control cells. These
results imply that there are conformational restraints on the DNA of
polyamine-depleted cells, a notion suggested by other studies as
well. It remains, however, an important future task to pinpoint the
specific roles of polyamines for chromatin structure.
In most
cell types, the biochemical characteristics of programmed cell death
include the activation of endogenous calcium and magnesium dependent
endonucleases, leading to fragmentation of the chromosomal DNA.
Previous results from our and other laboratories indicate that DNA
is destabilized in polyamine-depleted cells. This results in a
greater vulnerability of DNA to nucleasal attack. Because DNA is
known to become degraded during programmed cell death, we have begun
to investigate if polyamine degradation plays an active part in the
process. As one model system, we are using glucocorticoid-induced
programmed cell death in mouse
thymus. |
Return to
the top of this page
Return to
the Cell Proliferation Group research page
|


